Genetic Codes Explained
Haplotypes, recessives and genetic codes explained
This page is designed to help explain the current genetic codes which are found after the names of registered Holstein animals on pedigrees, web factsheets and top lists.
Firstly here are a few definitions of common genetic terms, which will be used in the description and explanation of genetic codes. The diagrams below can help visualise these genetic components. The further reading section at the end of this document also alerts you to some useful explanatory material on the subject.
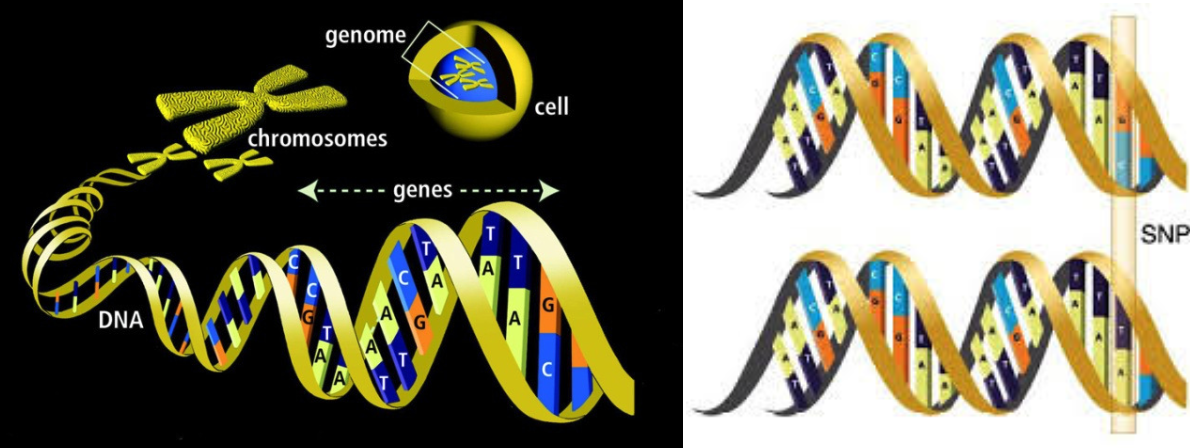
A Gene: is a sequence DNA which codes (i.e. is the instruction manual) for a particular amino acid. These amino acids are the building blocks of proteins. Each animal has two copies of each gene (which can vary - see Allele below).
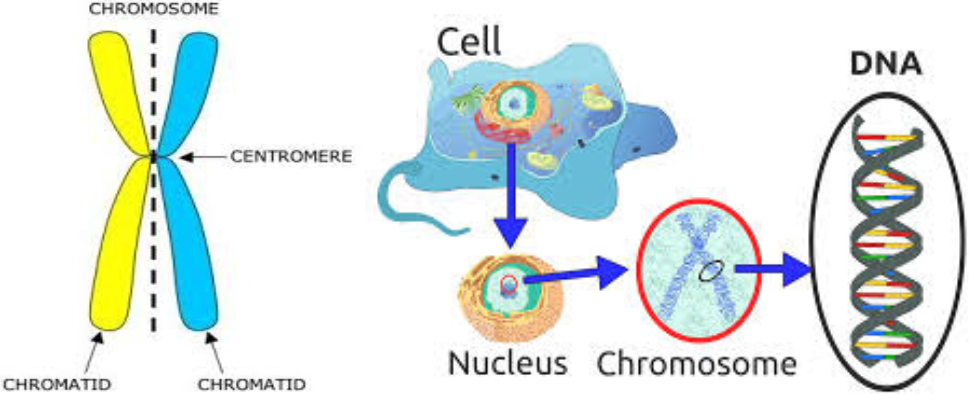
A Chromosome: is a large unit of DNA, containing many genes. Each chromosome is made up of a pair of chromatids. Cattle have 30 chromosomes found in most cells in the body. In Sperm and Eggs, Chromatids exist as un- paired single entities. Therefore an animal inherits half of each chromosome (and thus one copy of each gene) from each parent.
An Allele: is a form or version of a gene. An animal can have the same allele on both chromosomes (Homozygous) or different alleles (Heterozygous). Genes can have multiple alleles, such as the MCR1 gene responsible for some of the red coat colour variations, however 2 is most common. An animal can only inherit one allele from each parent.
A Recessive allele: is one which only has an effect when an animal inherits this version of the gene from each parent. A dominant allele affects an animal when either one or two versions are inherited. Most harmful alleles are recessive.
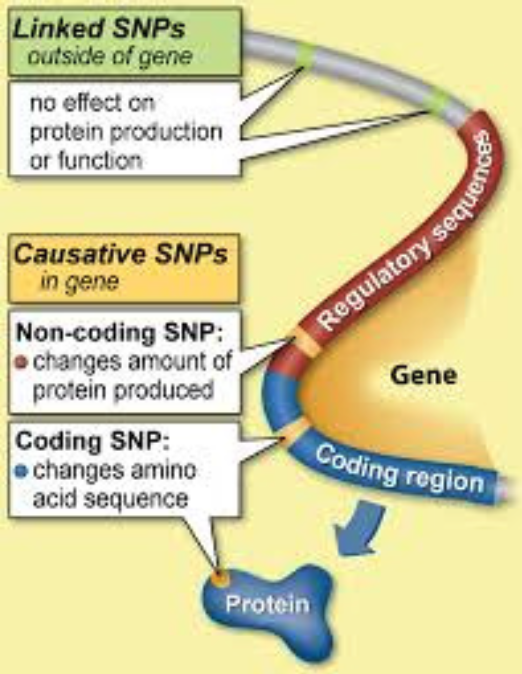
A SNP (Single Nucleotide Polymorphism): is a single unit of DNA which varies between animals within the same breed or species. A SNP may be located within a gene or on a region between genes on a chromosome. SNP are the markers used to genotype animals for genomic selection.
A Haplotype: is a group of SNP or alleles located close to each other on the chromosome, which are usually inherited together.
| Gene Name | Description | Gene and Expression Code |
| BLAD |
Bovine Leukocyte Adhesion Deficiency (deficiency of a normally occurring protein needed for white blood cells or leukocytes, which are body’s infection fighters) |
BLC = tested carrier of BLAD BLF = tested non-carrier of BLAD |
| Mule Foot |
Mule-Foot (toes of foot are joined, giving animal a single hoof, instead of cloven ones) |
MFC = tested carrier of Mule foot MFF = tested non-carrier of Mule foot |
| DUMPS |
Deficiency of Uridine Monophosphate Synthase (one of many enzymes contributing to normal metabolic processes) |
DPC = tested carrier of DUMPS DPF = tested non-carrier of DUMPS |
| CVM |
Complex Vertebral Malformation (causes still-born calves, abortions, and early embryonic losses) |
CVC = tested carrier of CVM CVF = tested non-carrier of CVM |
| Factor X1 |
Factor X1 (blood clotting disorder) |
XIC = tested carrier of Factor X1 XIF = tested non-carrier of Factor X1 |
| CIT |
Citrullinemia (accumulation of ammonia and other toxics in blood in young calves) |
CNC = tested carrier of Citrullinemia CNF = tested non-carrier of Citrullinemia |
| Brachyspina |
Brachyspina (causes abortion and stillborn, shortened spinal cord, long legs and abnormal organs) |
BYC = tested carrier of Brachyspina BYF = tested non-carrier of Brachyspina |
| Polled |
Animals without horns (reported born hornless - Not Tested) |
POR = code |
| Polled (Current Indirect Test) |
Indirect Test |
POS= tested true polled (homozygous PP) POC = tested carrier of polled (heterozygous Pp) POF= tested free of polled |
Red Alleles
| Coat Colour | Gene | Gene & Expression Code |
| RED |
Red gene (MCR1) |
RDC = carrier of red gene RDF = tested non-carrier of red gene |
| RED |
Variant Red gene |
VRR = not tested/determined by lineage. |
| BLACK/RED |
Black/red gene (MCR1) |
BRC = carrier of black / red gene |
| BLACK |
Black gene (MCR1) |
BKC = carrier of black gene |
Older Genetic Codes
Whilst the above tables show the codes currently in use for known alleles of interest, older codes were previously used for a number of these conditions. Ancestors of animals in Holstein UK factsheets can therefore display some of these older codes. CVM carriers were for example previously coded CV, rather than CVC as now adopted. Holstein UK now codes all animals following Word Holstein Friesian Federation (WHFF) recommendations. Some foreign Herdbooks also use different coding. Where animals are imported, or have semen imported and are thus are registered with Holstein UK, their codes will be recorded according to the WHFF format. When making breeding decisions based on genetic codes, it is key to look at the codes of the animals being mated, rather than those of their ancestors.
How do we test for genetic recessives?
Direct tests: are where a single ‘causative’ SNP is used. The genetic recessive is therefore either there or not and this is thus an accurate test.
Indirect tests: are predicted from groups of alleles on the chromosome, and depending upon the lab, different areas may be used and therefore different levels of accuracy attained.
It is therefore important to know whether a recessive test is direct or indirect as the level of accuracy will be affected.
Examples:
1. CVM is a directly tested genetic recessive. On the chromosome the causative SNP is either present or not. WHFF codes it as CVC (tested carrier) or CVF (tested non carrier).
2. Polled is an indirect test and the polled allele is dominant rather than recessive.
3. Brachyspina was originally an indirect test but there is now a direct test. In the Holstein UK we cannot discriminate between the two types of test, they are all marked as BYC or BYF.
Applying for a test
A Holstein UK member can submit a DNA sample for an animal and request a genetic recessive test for Brachyspina, CVM, BLAD, red factor and free martin. These genetic recessives can be divided into two groups; those that are directly predicted and those that are indirectly predicted.
Haplotypes affecting fertility
Haplotypes have been discovered which impact on the fertility of cattle. In order to avoid failed conceptions or early embryonic death, it is important that haplotypes and their negative effects are understood.
The term “haplotype” refers to a group of Alleles that are located at nearby positions on the chromosome which are usually inherited together.
Modern genomic tools have been used to identify thousands of haplotypes on each chromosome; each has a positive, neutral or negative association with production, conformation, health and fertility.
Five haplotypes have recently been discovered which negatively impact Holstein fertility. They are known as HH1, HH2, HH3, HH4 and HH5. Animals which are tested will be designated as either be carriers (C) or tested non-carriers (T).
The reasons why haplotypes impact fertility are unknown, however it is thought that inheritance of the same haplotype from each parent results in failed conception or early embryonic death.
Using semen from bulls with these haplotypes typically results in around 3% lower conception rate.
Haplotype inheritance
1. If both parents are carriers of an undesirable haplotype (HH1C):
There is a 25% chance that there will be an affected offspring that would not survive to birth
Of the live offspring, one-third will be unaffected non-carriers and two-thirds will be carriers, for example: HH1C Cow (carrier = Rr) x HH1C Cow (carrier = Rr) R = Normal haplotype
r = HH1 Haplotype (containing the causative mutation)
2. If the dam is unknown, but the grandsire and bull are both unaffected carriers of an undesirable haplotype (HH1C): There is a 12.5% chance that the resulting embryo will not survive to birth
3. If the cow and the bull were carriers of different haplotypes, e.g. if the cow was HH1C and the bull was HH2C, the following resulting offspring could be expected:
25% non-carriers of both (HH1T and HH2T)
25% carriers of one (HH1C)
25% carriers of the other (HH2C)
25% carriers of both (HH1C and HH2C)
NB. Fertility Haplotypes HH2 and HH3 are an indirect test, with prediction based on a group of SNPs. Recently the ‘causative’ SNP for HH1 has been discovered by USDA and is being used by some laboratories.
Breeding decisions and fertility haplotypes
It is desirable to avoid mating harmful haplotype carriers together. However, not using for example sires carrying these haplotypes is not necessarily practical. Firstly, many more undesirable haplotypes are likely to be found in the future. Secondly, these sires can carry other valuable economic traits. Therefore, breeders should not necessarily avoid bulls with these haplotypes nor cull cows, heifers and calves that are carriers. Rather, it is better to manage breeding decisions by avoiding mating known carriers.
How do I find out if my cattle are carriers?
Please contact member services on 01923 695223 or susanboughton@holstein-uk.org
Further Reading
WHFF
http://www.whff.info/info/geneticrecessives.php http://www.whff.info/info/documents/WHFFFutureassignedcodesforpolled_004.p df
Holstein USA
http://www.holsteinusa.com/pedigree_info/genetic_codes_traits.html Holstein USA – Interpreting and Utilising New Holstein Genetic Information http://www.holsteinusa.com/pdf/haplotype_details.pdf http://www.holsteinusa.com/pdf/haplotypes_affecting_fertility_080511.pdf http://www.holsteinusa.com/news/press_release2013.jsp#pr2013_20
